I2BMI¶
Biomedical Informatics toolkit by Institute for Informatics at Washington University School of Medicine in St. Louis
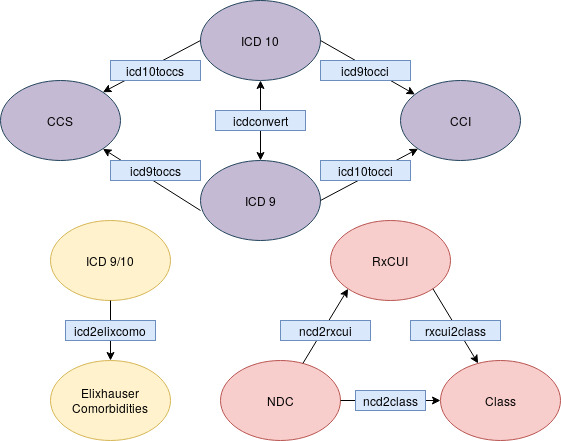
This package provides tools for converting medical data between different formats. Here is a diagram showing which function to use, based on the source and destination formats:
API Guide¶
-
i2bmi.elixcomoscore(df, col_icd, col_id)¶ score patients based on elixhauser comorbidities van Walraven C, Austin PC, Jennings A, Quan H, Forster AJ. A modification of the Elixhauser comorbidity measures into a point system for hospital death using administrative data. Medical care. 2009 Jun 1:626-33.
Parameters: - df – A Pandas DataFrame containing a column with patient IDs (col_id) and ICD 9 or 10 codes (col_icd).
- col_icd – The column containing ICD codes.
- col_id – The column containing patient IDs.
Returns: A Pandas DataFrame with three columns named col_id, “ElixComo”, and “ElixComoScore”. The ElixComo column stores particular comorbidities, while the ElixComoScore column stores the score for each comorbidity. The id column specified by the user will not be renamed.
-
i2bmi.icd10tocci(df, col_icd='icd10')¶ convert icd10 to CCI
Parameters: - df – A Pandas Dataframe containing ICD-10 data
- col_icd – Name of the column containing ICD-10 data
Returns: A dataframe with new columns for the CCI data corresponding to each ICD-10 code.
-
i2bmi.icd10toccs(df, col_icd='icd10')¶ convert icd10 to ccs
Parameters: - df – A Pandas Dataframe containing ICD-10 data
- col_icd – Name of the column containing ICD-10 data
Returns: A dataframe with new columns for the CCS data corresponding to each ICD-10 code.
-
i2bmi.icd9tocci(df, col_icd='icd9')¶ convert icd9 to CCI
Parameters: - df – A Pandas Dataframe containing ICD-9 data
- col_icd – Name of the column containing ICD-9 data
Returns: A dataframe with new columns for the CCI data corresponding to each ICD-9 code.
-
i2bmi.icd9toccs(df, col_icd='icd9')¶ convert icd9 to CCS
Parameters: - df – A Pandas Dataframe containing ICD-9 data
- col_icd – Name of the column containing ICD-9 data
Returns: A dataframe with new columns for the CCS data corresponding to each ICD-9 code.
-
i2bmi.icdconvert(df, col_icd='icd', icd_version=9)¶ convert icd 9 to 10, or 10 to 9 https://www.cms.gov/Medicare/Coding/ICD10/2018-ICD-10-CM-and-GEMs.html
Parameters: - df – Pandas DataFrame containing ICD 9 or 10 data
- col_icd – Name of the column in df with ICD codes
- icd_version – The source version.
Returns: A pandas Dataframe with converted ICD codes.
-
i2bmi.icdtoelixcomo(df, col_icd)¶ convert icd 9 or 10 to Elixhauser Comorbidities
Parameters: - df – A Pandas DataFrame containing ICD 9 or 10 data.
- col_icd – Name of the column in df containing ICD 9 or 10 codes.
Returns: A Dataframe with two new columns, ElixComo (containing Elixhauser comorbidities) and ElixComoScore (containing the scores for each comorbidity).
-
i2bmi.load_cci10()¶ load mapping file from icd10 to Chronic Condition Indicator (CCI) https://www.hcup-us.ahrq.gov/toolssoftware/chronic_icd10/chronic_icd10.jsp
Returns: Mapping file from icd10 to Chronic Condition Indicator (CCI)
-
i2bmi.load_cci9()¶ load mapping file from icd9 to Chronic Condition Indicator (CCI) https://www.hcup-us.ahrq.gov/toolssoftware/chronic/chronic.jsp
Returns: Mapping file from icd9 to Chronic Condition Indicator (CCI)
-
i2bmi.load_ccs10()¶ load mapping file from icd10 to Clinical Classification Software (CCS) https://www.hcup-us.ahrq.gov/toolssoftware/ccs10/ccs10.jsp
Returns: Mapping file form icd10 to Clinical Classification Software (CCS)
-
i2bmi.load_ccs9()¶ load mapping file from icd9 to Clinical Classification Software (CCS) https://www.hcup-us.ahrq.gov/toolssoftware/ccs/ccs.jsp
Returns: Mapping file from icd9 to Clinical Classification Software (CCS)
-
i2bmi.load_icd10to9()¶ load icd 10 to icd 9 mapping file https://www.cms.gov/Medicare/Coding/ICD10/2018-ICD-10-CM-and-GEMs.html
Returns: A file mapping icd 10 to icd 9
-
i2bmi.load_icd9to10()¶ load icd 9 to icd 10 mapping file https://www.cms.gov/Medicare/Coding/ICD10/2018-ICD-10-CM-and-GEMs.html
Returns: A file mapping icd 9 to icd 10
-
i2bmi.loadelixcomo()¶ load Elixhauser comorbidities mapping file Quan H, Sundararajan V, Halfon P, Fong A, Burnand B, Luthi JC, Saunders LD, Beck CA, Feasby TE, Ghali WA. Coding algorithms for defining comorbidities in ICD-9-CM and ICD-10 administrative data. Medical care. 2005 Nov 1:1130-9.
Returns: The Elixhauser comorbidities mapping file
-
i2bmi.ndc2class(df_med, col_ndc='ndc', getname=True, indexcol='ROWID', timecol='TIME')¶ combine NDC2RXCUI and RXCUI2CLASS to go from NDC to CLASS
Parameters: - df_med – A Pandas DataFrame with a column containing ndc data
- col_ncd – Name of the column in df_containing NDC data
- getname – If True, this function will internally use the class name as the identifier. If False, this function will internally use the class ID.
- indexcol –
- timecol –
Returns: A Pandas DataFrame with additional drug class data.
-
i2bmi.ndc2rxcui(df_med, col_ndc='ndc')¶ convert NDC codes to RXCUI
Parameters: - df_med – A Pandas DataFrame with a column containing ndc data
- col_ncd – Name of the column containing NDC data
Returns: A Pandas DataFrame with the following columns: ndc (the NDC identifier), rxcui (the corresponding RXCUI identifier), start (the start date), and end (the end date).
-
i2bmi.onehotify(df, col_id, col_val)¶ onehotifying categorical columns
-
i2bmi.parsediag(dfin, col_icd, col_id, icd_version)¶ parse diagnosis dataset and include Clinical Classification Software, Chronic Glag, and Elixhauser Comorbidity
-
i2bmi.rxcui2class(df_mapin, getname=True)¶ convert RXCUI to drug classes
Parameters: - df_mapin – A Pandas DataFrame containing RXCUI identifiers. This must be present in a column called “rxcui”.
- getname – If True, this function will internally use the class name as the identifier. If False, this function will internally use the class ID.
Returns: A Pandas DataFrame with additional drug class data.